Our laboratory focuses on research and development of novel device technologies with the aim of applying nano- and micrometer-sized fabrication techniques to the medical and biological fields. Below are some of the research topics we have been working on recently.
Mechanical Processing of Biosamples
Tissue/Cell Spatial Dissection
In medicine and biology, a number of technologies have been developed to analyze the types and amounts of biomolecules contained in single cells. On the other hand, spatial information on the location of the analyzed cells in the body and the organ from which they originated is often lost.
We are working on the development of a technology to sample cell-sized fractions with spatial information, that is, with the tag of where they originated from in an organ. This technology will make it possible to link the material information of analyzed cells with spatial information in organs, and is expected to lead to the analysis of cancer microstructures and the elucidation of the relationship between cytoarchitecture and function.
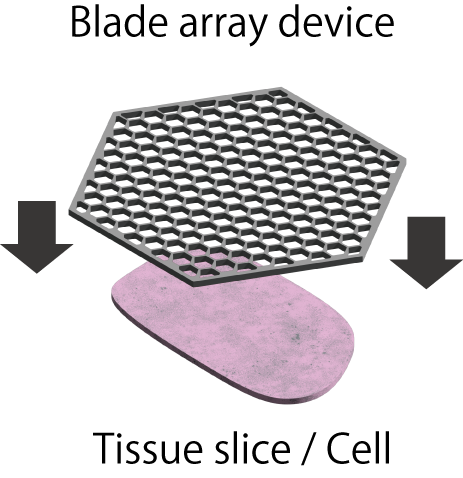
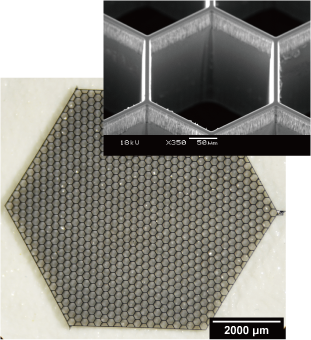
We have developed a device named “blade array device,” which is composed of an array of sharp blade-like structures using microfabrication technology, and are aiming to realize cell sampling with spatial information. Using this device, we are developing a new cell processing technology called “spatial dissection”, in which a single cell is divided into multiple micro-regions or an organ is divided into single cell-sized sections by physical cutting.
Ref.
- (Japanese) https://wakasapo.nedo.go.jp/seeds/seeds-4720/
- (Japanese) https://www.youtube.com/watch?v=Op3y6Dx97lk
Optically-Driven Nanotools
At the focus of a laser beam, tiny objects with sizes ranging from nanometers to micrometers can be captured, which is known as optical tweezers. Our goal is to create various shapes of micro objects captured by optical tweezers and to apply surface treatment to them in order to harness the functions and use them as tools in the nano-world. By using such optically-driven nano/microtools, we are approaching areas that are difficult to handle in conventional bio experiments, such as manipulation and processing of a single cell and a single biomolecule.
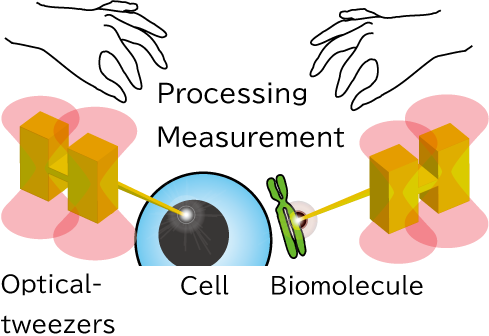

The optically-driven tools we develop can be fabricated in large batches using semiconductor microfabrication technology, and can be obtained in a million pieces in aqueous solution. The tools can be designed in various shapes and can be functionalized by forming metal films on their surfaces or by immobilizing enzymes or antibodies through surface modification.
Ref.
- M. Harada, H. Takao, F. Shimokawa, K. Terao*: “Development of optically driven nanoneedles using SU-8 nanofabrication”, Electronics and Communications in Japan, 105(1), e12327 (2022)
- K. Terao*, C. Masuda, R. Inukai, M. Gel, H. Oana, M. Washizu, T. Suzuki, H. Takao, F. Shimokawa, F. Oohira: “Characterization of optically-driven microstructures for manipulating single DNA molecules under a fluorescence microscope”, IET Nanobiotechnology, 10(3), 124-128(2016)
Assembly of Organs from Single Cells
In the fields of drug discovery and regenerative medicine, it is expected that organ structures can be formed in vitro to replace animal experiments, or to be used in the preliminary stages for clinical trials as samples that are closer to in vivo conditions than normal cultured cells. Methods of self-assembling cell clusters and bioprinting have been proposed, but it is difficult to control the type and position of the cells at the single-cell level. To solve this problem, we aim to assemble individual cells as desired and form various cellular tissue structures.
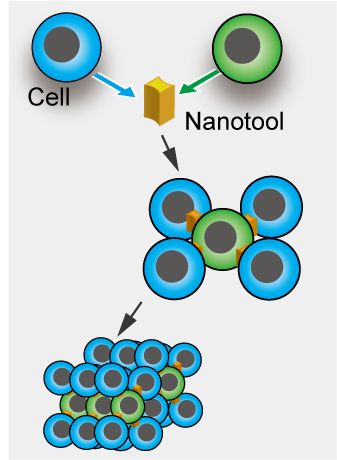
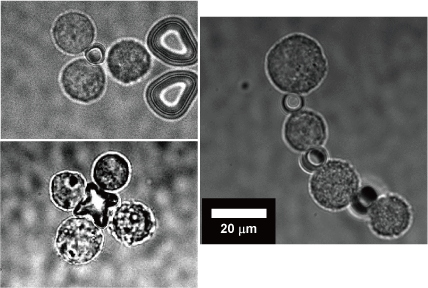
We have applied the optically-driven nanotools to cell assembly. By modifying the surface of the structures fabricated by semiconductor microfabrication technology with molecules that adhere to cells, the tool functions as an adhesive to connect cells together. By manipulating this tool and cells with laser beam, cells can be assembled like blocks in a short time.
Ref.
- S. Mori, T. Ito, H. Takao, F. Shimokawa, K. Terao*: “Optically-driven microtools with an antibody-immobilized surface for on-site cell assembly”, IET Nanobiotechnology, 1-7 (2023)
Single Molecule Mechanical Processing
Biomolecules are usually processed as a mass of molecules in aqueous solution. Therefore, information on individual molecules is lost and averaged. In addition, DNA molecules, a typical biomolecule, are linear polymers, but they are easily fragmented by the sheer flow of solution, making it difficult to treat the molecules intact. Therefore, most of the widely used biomolecular analysis techniques target molecular populations, and for DNA molecules, analysis is performed by amplifying the fragmented DNA molecules. In this process, differences between individual molecules, spatial structure and function are lost. Therefore, we are developing technologies to manipulate, process, and analyze molecules on a molecule-by-molecule manner, rather than on a collective.
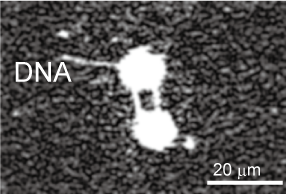
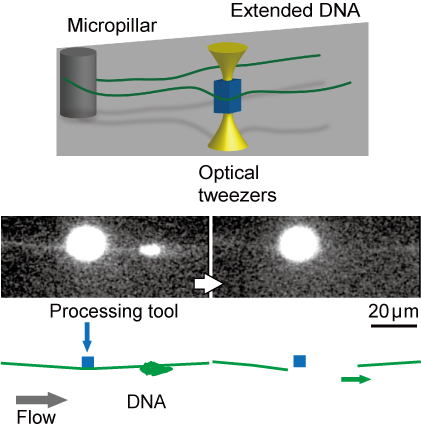
In order to perform various observation experiments using single DNA molecules under a microscope, we have developed techniques for manipulating the morphology of DNA molecules using lasers and electro-osmotic flow, and for cutting and recovering target DNA molecules at targeted positions to enable recovery of specific sites of DNA with spatial information.
Ref.
- A. Masuda, H. Takao, F. Shimokawa, K. Terao*: “On-site processing of single chromosomal DNA molecules using optically driven microtools on a microfluidic workbench”, Scientific Reports, 11, 7961 1-9 (2021)
- R. Inukai, H. Takao, F. Shimokawa, K. Terao*: “Capture and elongation of single chromosomal DNA molecules using optically-driven microchopsticks”, Biomicrofluidics, 14, 044114 (2020)
- K. Terao*, M. Washizu, H. Oana: “On-Site Manipulation of Single Chromosomal DNA Molecules by using Optically Driven Microstructures”, Lab on a Chip, 8(8), 1280-4 (2008)
Single Cell Stimulation and Response Analysis
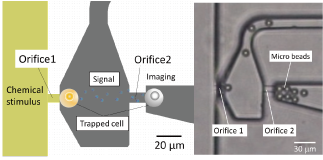
Local Drug Stimulation to Single Cell for Receptor Analysis
In drug discovery and cell biology, it is a fundamental experiment to measure the cellular response of cells to a drug. Many drugs target receptors exist on the cell surface. Receptors interact with drug molecules to induce or inhibit cellular responses. The relationship between the number of receptors that interact with a drug and the amount of cellular response is thought to be different for each cell type and cell state, but is not well understood. In this research, we are focusing on this point and developing a technology to analyze the relationship between drug-receptor interactions and cellular responses on single cell resolution using microfluidic devices. This technology will contribute to drug discovery and personalized medicine by clarifying the response of each cell.
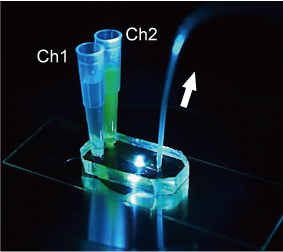
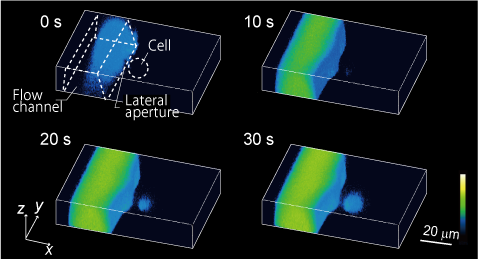
We have developed microfluidic devices and are developing new experimental techniques to measure cellular responses to local stimuli and the localization and propagation of signals by applying drugs to a very small defined area of the cell surface. Through these efforts, we are currently developing methods to quantitatively analyze the relationship between receptors and cellular responses with single cell resolution.
Ref.
- K. Terao*, M. Gel, A. Okonogi, A. Fuke, T. Okitsu, T. Tada, T. Suzuki, S. Nagamatsu, M. Washizu, H. Kotera: “Subcellular Glucose Exposure Biases the Spatial Distribution of Insulin Granules in Single Pancreatic Beta Cells”, Scientific Reports, 4, 4123 1-6 (2014)
- K. Terao*, A. Okonogi, A. Fuke, T. Okitsu, T. Suzuki, M. Washizu, H. Kotera: “Localized substance delivery to single cell and 4D imaging of its uptake using a flow channel with a lateral aperture”, Microfluidics and Nanofluidics, 12(1), 423-429 (2012)
Drug Stimulation to iPS Embryoid Body for Differentiation Control
The process of transforming iPS cells into target cells is called differentiation induction. Differentiation induction is performed using drugs, etc., but cells that remain undifferentiated or are not the target cells are generated, resulting in a heterogeneous cell population even when the same differentiation induction procedure is performed. This heterogeneity in differentiation induction is one of the hurdles to the practical use of iPS cells from the viewpoint of the risk of tumor formation and cell quality assurance.
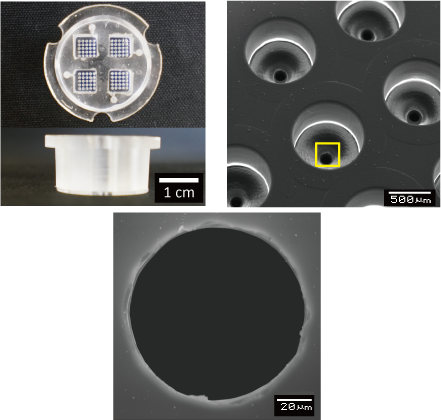
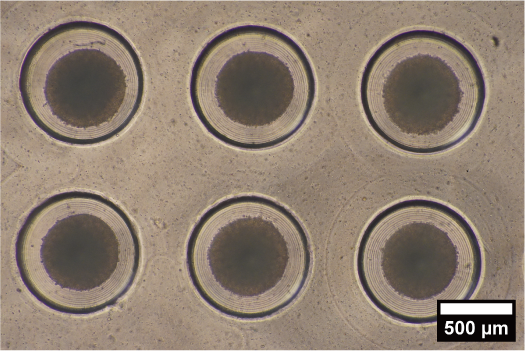
We are attempting to control differentiation induction by forming embryoid bodies with microdevices that can reproduce an environment similar to that in vivo, and by regulating the microenvironment around the embryoid body. By developing novel microdevices that adapt to the widely used dish and well plate standards, we ensure compatibility with a variety of liquid handling and analysis devices.
Ref.
- N. Kusunoki, S. Konagaya, M. Nishida, S. Sato, H. Takao, F. Shimokawa, K. Terao*: “Microfluidic device for differentiation induction of iPS cells with local chemical stimulation”, Electronics and Communications in Japan, 105(3), e12393 (2023)
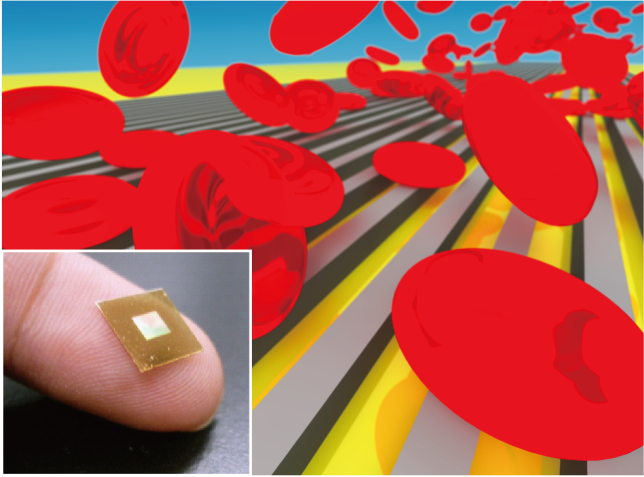
In Vitro Model for Analyzing Blood Circulation of Cancer Cells
Most cancer deaths are accompanied by metastasis, a process in which cancer cells circulate in the bloodstream. It is extremely difficult to observe this process in vivo due to the rarity and rapid move of cancer cells in the blood. Therefore, technologies have been proposed to reproduce the blood circulation process of cancer cells in vitro. In particular, considerable attention has been paid to the behavior of cancer cells in capillaries, which are thought to have a significant impact on metastasis, and how cancer cells deform and pass through constrictions and how they recover and change their properties once they pass through remain unsolved.
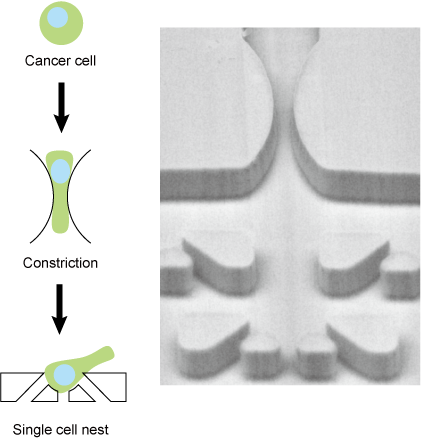
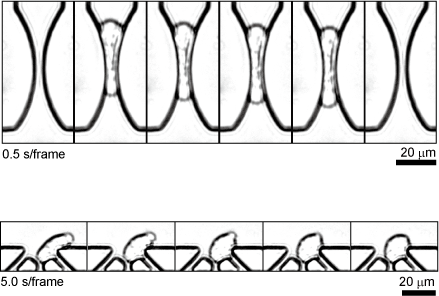
The microfluidic device we have developed not only reproduces the micro constrictions of blood capillaries, but also has micro nest structures for capturing and observing cancer cells immediately after they deform at constriction structures. This is expected to lead to the analysis of cellular events that occur during the blood circulation of cancer cells and, in the future, to the diagnosis of clinical cancer cells.
Ref.
- E. Gasser*, E. Su, K. Vaidziulyte, N. Abbade, H. Cognart, J.B. Manneville, J.L. Viovy, M. Piel, J.Y. Pierga, K. Terao*, C. Villard*: “Deformation under flow and morphological recovery of cancer cells”
Lab on a Chip, 24, 3930-3944 (2024)
Biosample Manipulation
Microfluidic Device Reproducing Kidney Microenvironment
We are developing a microfluidic device to reproduce a kidney microenvironment in vitro. In particular, we aim to reproduce the conditions of septic acute kidney injury (AKI) to understand disease mechanisms and to develop effective therapies. However, it is difficult to observe them in vivo.
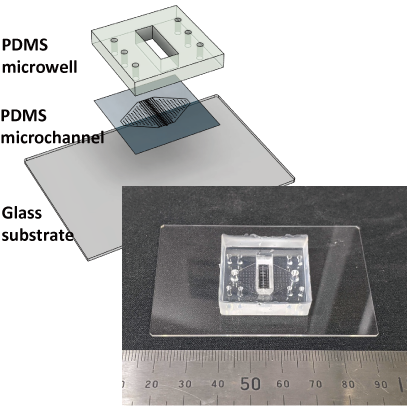
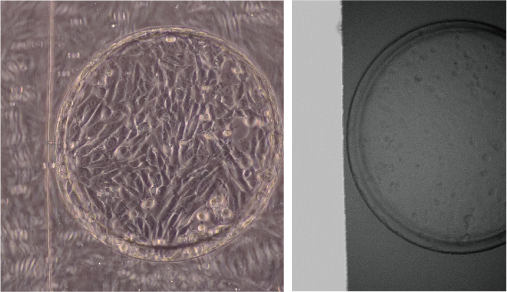
Therefore, we are developing a device that can reproduce the passage of urine in vitro (renal tubule) in microfluidic channels and observe changes in tubular cells at the single-cell level during septic shock, as well as measure urine leakage and interactions with other types of cells.
“Molecular Ring Toss” for Imaging Single Circular DNAs
Conformational changes in DNA molecules play an important role in the maintenance of living functions and diseases through the expression and repression of functions in cells. For this reason, many methods have been developed to measure the dynamics of single DNA molecules. However, most of them are limited to DNA molecules on a linear strand with ends. In contrast, we focused on circular DNA molecules. Since circular DNA molecules do not have ends, they are suitable for observing specific conformational changes such as molecular twisting and relaxation
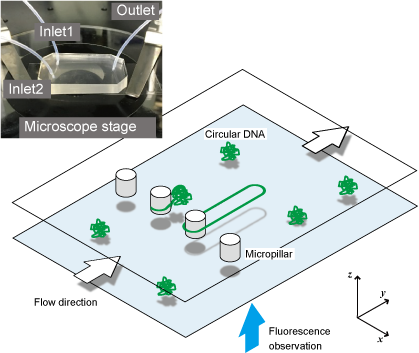
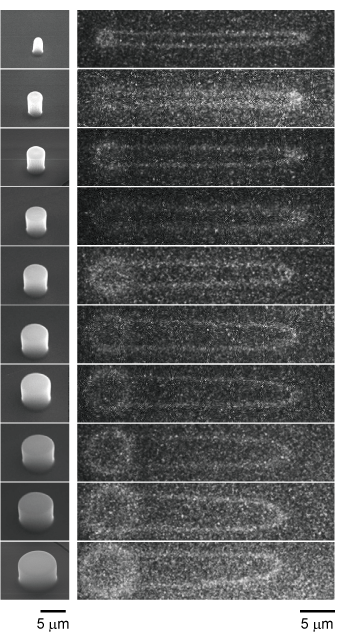
Chemical modifications to DNA ends widely used for single molecule manipulation are not available for circular DNA. Therefore, we have developed a technique to physically trap circular DNA molecules and visualize them under a microscope. By hooking circular DNAs onto the micropillar structures like a “ring toss” and extending them by flow, the dynamics of circular DNA can be observed with a single molecule under a fluorescence microscope.
Ref.
- D. Dohi, K. Hirano*, K. Terao*: “Molecular ring toss of circular BAC DNA using micropillar array for single molecule studies”, Biomicrofluidics, 14, 014115 (2020)
2-Dimentional Micronozzle Array
A micro-nozzle that dispenses solution through a tiny aperture can be used to fabricate fine fibers and perform chemical processing on a limited area of a substrate surface. For this reason, they are used in bioprinting and biopatterning. If these micro-nozzles can be densely integrated in a two-dimensional array, it is expected that complex flows of various solutions can be formed from the apertures, leading to the in vitro formation of organs and highly integrated biosensors.

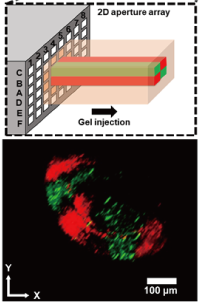
We have developed high-density micro-nozzle arrays by stainless steel microfabrication and resin molding. Using this device, we have performed formation of heterogeneous gel fibers with adjustable cross-sectional patterns and simultaneous micro-patterning of multiple antibodies on the surface of substrates.
Ref.
- K. Takahashi, H. Takao, F. Shimokawa, K. Terao*: “On-demand formation of heterogeneous gel fibers using two-dimensional micronozzle array”, Microfluidics and Nanofluidics, 26, 15 (2022)
- K. Takahashi, S. Kamiya, H. Takao, F. Shimokawa, K. Terao*: “Stainless Microfluidic Probe with 2D array Microapertures”, AIP Advances, 11(1), 015331 (2021)
Previous Works
Surface Biosensing Enhanced by Microstructures
In medical diagnosis and drug discovery, there is a growing demand for sensing biomolecules with low molecular weight and low concentration. For example, it is expected to realize early diagnosis of cancer by detecting cancer markers in blood. We have focused on surface biosensors, which have the advantage of non-labeled sensing, and have worked to increase sensitivity and functionality using MEMS technology with the aim of applying them to medical, food, and basic research applications.
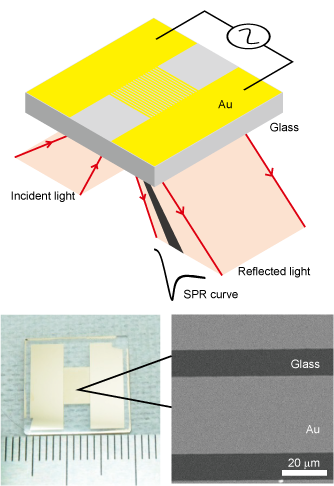
We have developed a filter SPR sensor chip integrated with size discrimination function by fabricating a micro-slit structure on a surface plasmon resonance (SPR) sensor chip. It is capable of measuring aggregation and de-aggregation of nanoparticles and can detect blood proteins in less than 100 seconds from whole blood.
Other developments include a gold-black biosensor chip with a nanometer-sized gold structure (black gold) formed by plating on the biosensor surface, and a comb-shape electrode sensor chip capable of applying an alternating electric field. Several-fold improvement in detection was obtained in the quartz crystal microbalance method (QCM method) and the surface plasmon resonance method (SPR method).
Ref.
- K. Terao*, S. Kondo: “AC-Electroosmosis-Assisted Surface Plasmon Resonance Sensing for Enhancing Protein Signals with a Simple Kretschmann Configuration”, Sensors, 22(3), 854 1-9 (2022)
- K. Terao*, S. Hiramatsu, T. Suzuki, H. Takao, F. Shimokawa, F. Oohira: “Fast protein detection from raw blood by size-exclusion SPR sensing”, Analytical Methods, 7, 6483-6488 (2015)
- N. Nagase, K. Terao*, N. Miyanishi, N. Tamai, N. Uchiyama, T. Suzuki, H. Takao, F. Shimokawa, F. Oohira: “Signal Enhancement of Protein Binding by Electrodeposited Gold Nanostructures for Application in Kretschmann-Type SPR Sensor”, Analyst, 137(21), 5034-5040 (2012)
- K. Terao*, K. Shimizu, N. Miyanishi, S. Shimamoto, T. Suzuki, H. Takao, F. Oohira: “Size-Exclusion SPR Sensor Chip: Application to Detection of Aggregation and Disaggregation of Biological Particles”, Analyst, 137(9), 2192-2198 (2012)
Single Cell Manipulation for Cell-Cell Interaction Studies
Cells function differently when they exist singly or in groups, and thus, cell-cell interactions play an important role in the expression of functions. Techniques have been developed to investigate cell-cell interactions and cellular functions by regulating the type and arrangement of cells in vitro using microdevices. We have developed microfluidic devices that can control the arrangement of cells at the single-cell level.
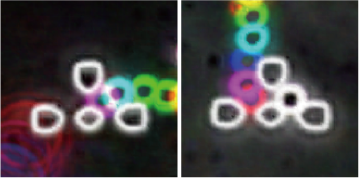
Because of the random positioning of cells in conventional culture dishes, it is difficult to measure interactions in a controlled environment. In contrast, we have developed a device that uses a flow driven by an electric field to analyze contact interactions between different cell types by placing two types of cells next to each other. We have also developed a device that analyzes non-contact interactions (paracrine) by placing two cells at a specified distance from each other and applying drug stimulation to only one of the cells.
Ref.
- Y. Tao, K. Fukuda, H. Takao, F. Shimokawa, and K. Terao*: “Development of microfluidic device for imaging paracrine communication”, IEEJ Transactions on Sensors and Micromachines, 137(5), 128-133(2017)
- K. Terao*, Y. Kitazawa, R. Yokokawa, A. Okonogi, H. Kotera: “Open-Access and Multi-Directional Electroosmotic Flow Chip for Positioning Heterotypic Cells”, Lab on a Chip, 11(8), 1507-1512(2011)
